MindTheGap performs detection and assembly of DNA insertion variants, whatever their size, in NGS read datasets with respect to a reference genome. Here are some results obtained on simulated data.
Some of these results were already shown in the initial publication (August 2014) below, but since the publication the software was greatly improved in terms of running time but also in terms of results quality (version 2.0.0). The main algorithmic improvement is to detect additionnal variants, such as SNPs and deletions. This feature improves the sensitivity of the insertion detection algorithm for insertions that are located near these other variants.
Publication
Guillaume Rizk, Anaïs Gouin, Rayan Chikhi and Claire Lemaitre. (2014) MindTheGap: integrated detection and assembly of short and long insertions. Bioinformatics, 30(24):3451-3457.
Simulations
Data and simulations are fully described in the publication. Briefly, artificial reads are simulated on the genome of C. elegans (2×100 bp paired-end, 40X). Some random portions of the genome are then deleted to produce a new reference genome, these deletions in the reference genome appear then as insertions in the read datasets. 2 insertion size ranges are shown here : 1-100 bp and 1000 bp. MindTheGap is then compared to SOAPindel.
Additionnally, we generated another reference genome with the same deletions as previously but containing also around 100,000 point mutations uniformly distributed along the chromosomes (1 every 1000 bp).
Results
Below are the results obtained for the artificial reference genome without point mutations. In these simulations, only insertion variants differenciate the reads from the reference genome. Therefore we expect similar results between the former and newer version of MindTheGap in terms of recall and precision. Nevertheless, in the new version, each prediction is now associated to a quality score, this enables to filter out the results to obtain a high precision subset (see the line MindTheGap v2.0.0 qual>30).
| Insertion size between 1-100 bp | Insertion size 1000 bp | |||||||
| Recall | Precision | Time* | Memory | Recall | Precision | Time* | Memory | |
| MindTheGap (published) | 94.2 | 99.2 | 36 min | 485 MB | 72.1 | 96.8 | 40 min | 481 MB |
| MindTheGap v2.0.0 | 94.2 | 98.9 | 13 min | 2 GB | 72.7 | 95.7 | 17 min | 2 GB |
| MindTheGap v2.0.0 qual>30 | 84.0 | 99.8 | 13 min | 2 GB | 63.0 | 98.9 | 17 min | 2 GB |
| SOAPindel | 92.2 | 99.3 | 60 min | 4.2 GB | 0.0 | 0.0 | — | — |
*: using 8 threads. Note: SOAPindel can not detect insertions larger than 200 bp (for this sequencing data).
Below are the results obtained with the dataset containing artificial insertions and SNPs. In this dataset, we expect that some SNPs can affect the detection of the artificial insertions.
| Insertion size between 1-100 bp | Insertion size 1000 bp | |||||||
| Recall | Precision | Time | Memory | Recall | Precision | Time | Memory | |
| MindTheGap (published) | 88.2 | 94.0 | 54 min | 480 MB | 66.6 | 71.8 | 45 min | 481 MB |
| MindTheGap v2.0.0 | 92.2 | 65.1 | 16 min | 2 GB | 71.2 | 71.9 | 20 min | 2 GB |
| MindTheGap v2.0.0 qual>30 | 81.6 | 99.5 | 16 min | 2 GB | 60.6 | 98.7 | 20 min | 2 GB |
| SOAPindel | 91.2 | 98.3 | 95 min | 4 GB | 0.0 | 0.0 | — | — |
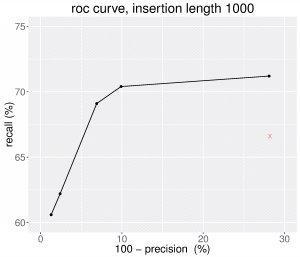
Here is the Precision-recall curve obtained with MindTheGap version 2.0.0 when we vary the quality score threshold (for insertions of size 1000 bp). We can see that the novel version of MindTheGap obtains better results in terms of False Negative and False Positive rates than the previous one (denoted by the red cross), and that the quality score enables to choose between better recall or high precision prediction sets.